Project: Cancer Cell Tracking
Published:
This project focuses on cell tracking based on Li et al.. By utilizing watershed algorithm and feature vectors (based on position, shape, spatial distribution and texture), the algorithm can succeed tracking cancer cells in image sequence with segmentation accuracy and tracking accuracy 69.51% and 76.78%, repectively.
Usage
The data can be found at Cell Tracking Challenge Website.
ipython notebook: to better show the algorithm step by step, besides the python scipts, I also create a ipython notebook to visualize the interim results.
Some explanation of the scripts:
main.py: the main procedure including all steps.
adaptivethresh.py: compute adaptive thresholding of image sequence in order to generate binary image for Nuclei segmentation.
gvf.py: compute gradient vector field (GVF) to find the seeds for following watershed.
watershed.py: segment cells
graph_construction.py: generate a neighboring graph contraction using Delaunary Triangulation.
matching.py: calculate feature vector for each cell and match cells.
Results
- Result of original image sequence.
- Result of tracking all cells.
- Result of tracking specific cell in mitosis.
- Plot of the previous tracking.
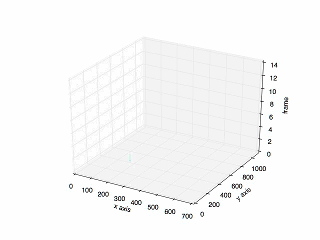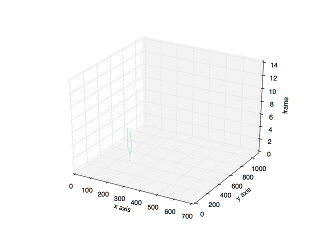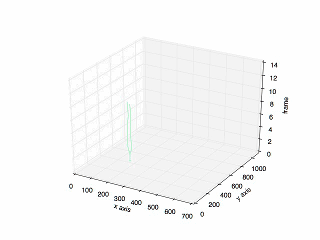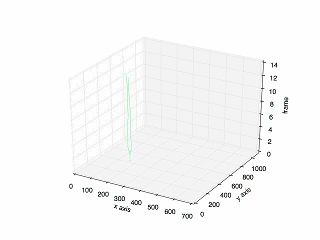
Looking for code? Here is the link to the repository in my Github.




Leave a Comment